#> `geom_smooth()` using formula = 'y ~ x'Simple
Logistic Regression
R Packages
- rcistats
- tidyverse
Heart Disease Data
Variables of Interest
age: Age of patientdisease: Indicating if they have heart disease

Modeling Binary Outcomes
Modeling Binary Outcomes
Logistic Regression
Logistic Regression in R
Prediction with Models
Modeling Binary Outcomes
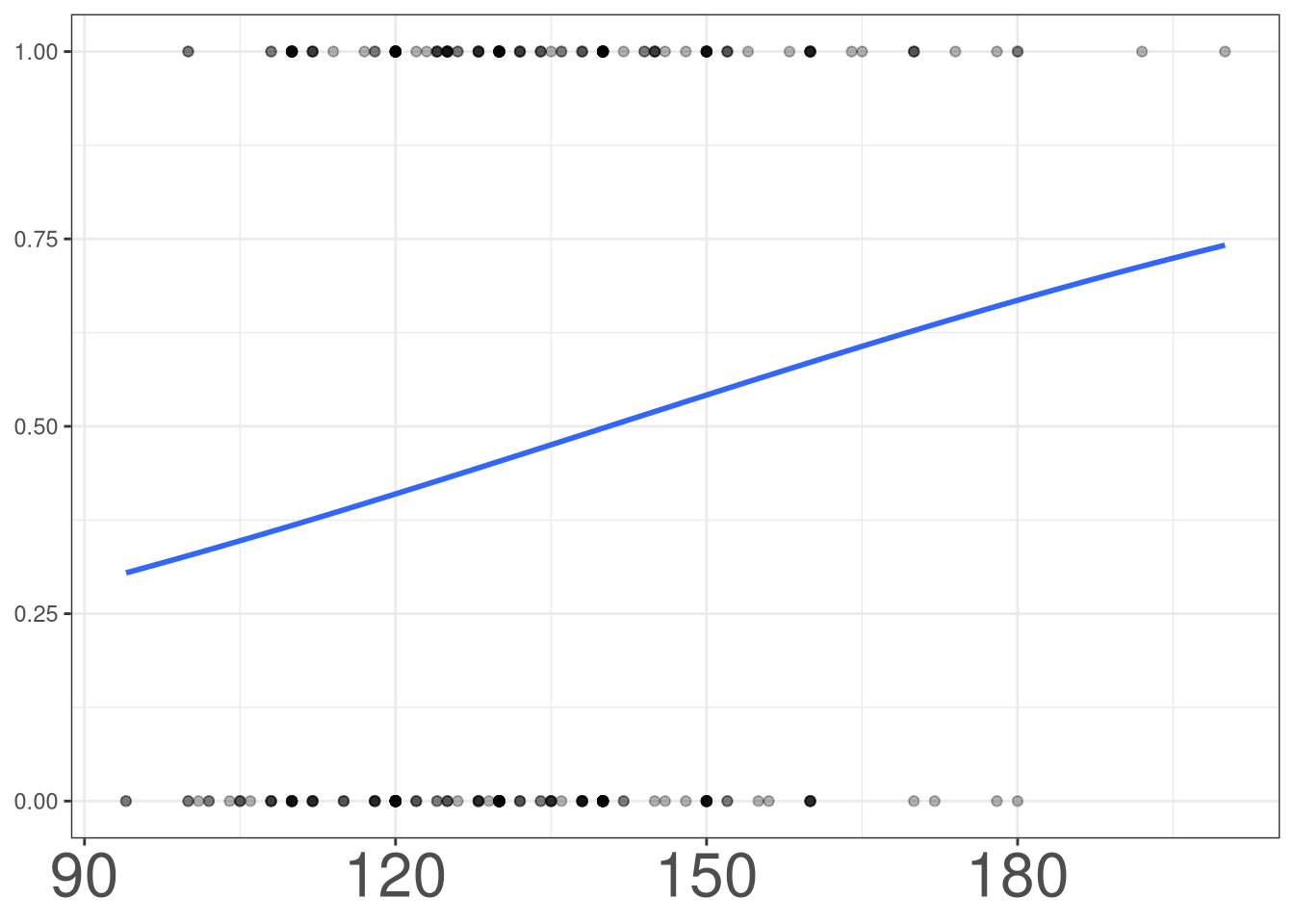
Modeling Binary Outcomes
#> `geom_smooth()` using formula = 'y ~ x'
Mathematical Model
\[ \left(\begin{array}{c} Yes \\ No \end{array}\right) = \beta_0 + \beta_1X \]
Let …
\[ Y = \left\{\begin{array}{cc} 1 & Yes \\ 0 & No \end{array}\right. \]
Construct a Model
\[ P\left(Y = 1\right) = \beta_0 + \beta_1X \]
Construct a Model
\[ P\left(Y = 1\right) = \frac{e^{\beta_0 + \beta_1X}}{1 + e^{\beta_0 + \beta_1X}} \]
Construct a Model
\[ \frac{P(Y = 1)}{P(Y = 0)} = e^{\beta_0 + \beta_1X} \]
where \(\frac{P(Y = 1)}{P(Y = 0)}\) are considered the odds of observing \(Y = 1\).
The Logistic Model
\[ \log\left\{\frac{P(Y = 1)}{P(Y = 0)}\right\} = \beta_0 + \beta_1X \]
Notation
\[ \log\left\{\frac{P(Y = 1)}{P(Y = 0)}\right\} = \log\left\{odds\ of \ 1\right\} = lo(1) \]
Logistic Regression
Modeling Binary Outcomes
Logistic Regression
Logistic Regression in R
Prediction with Models
Logistic Regression
Logistic Regression is used to model the association between a predictor and a binary outcome variable.
This is similar Linear Regression which models the association between a predictor and a numerical outcome variable.
Logistic Regression
Logistic Regression uses the logistic model to formulate the relationship between a predictor and the outcome.
More specifically, for an outcome of Y:
\[ Y = \left\{\begin{array}{cc} 1 & \text{Category 1} \\ 0 & \text{Category 2} \end{array}\right. \]
The predictor variable will model the probability of observing category 1 (\(P(Y=1)\))
Logistic Model
\[ \log\left\{\frac{P(Y = 1)}{P(Y = 0)}\right\} = \beta_0 + \beta_1X \]
Regression Coefficients \(\beta\)
The regression coefficients quantify how a specific predictor changes the odds of observing the first category of the outcome (\(Y = 1\))
Estimating \(\beta\)
To obtain the numerical value for \(\beta\), denoted as \(\hat \beta\), we will be finding the values of \(\hat \beta\) that maximizes the likelihood function:
\[ L(\boldsymbol \beta) = \prod_{i=1}^n \left(\frac{e^{\beta_0 + \beta_1X}}{1 + e^{\beta_0 + \beta_1X}}\right)^{Y_i}\left(\frac{1}{1 + e^{\beta_0 + \beta_1X}}\right)^{1-Y_i} \]
The likelihood function can be thought as the probability of observing the entire data set. Therefore, we want to choose the values the \(\beta_0\) and \(\beta_1\) that will result in the highest probability of observing the data.
Estimated Parameters
The values you obtain (\(\hat \beta\)) tell you the relationship between the a predictor variable and the log odds of observing the first category of the outcome \(Y=1\).
Exponentiating the estimate (\(e^{\hat \beta}\)) will give you the relationship between a predictor variable and the odds of observing the first category of the outcome \(Y=1\).
Interpreting \(\hat \beta\)
For a continuous predictor variable:
As X increases by 1 unit, the odds of observing the first category (\(Y = 1\)) increases by a factor of \(e^{\hat\beta}\).
Logistic Regression in R
Modeling Binary Outcomes
Logistic Regression
Logistic Regression in R
Prediction with Models
Logistic Regression in R
DATA: Name of the data frameY: Outcome Variable of Interest in the data frameDATAX: Predictor Variable in the data frameDATA
Model Information
The model_info function can provide basic information about a linear or logistic regression model.
It can provide what is being modeled (yes, 1) in logistic regression.
It can provide basic info of the predictor variables.
Model Information in R
MODEL: fitted model fromlm()orglm
Example
Modelling disease by age:
Code
#> Outcome Variable: disease#> Modeling Probability: yes#> Numerical Predictors:
#> age#> Categorical Predictors:
#> cp:
#> Asymptomatic (Reference)
#> Non-anginal Pain
#> Atypical Angina
#> Typical Angina
#> sex:
#> Female (Reference)
#> Male
#> restecg:
#> Normal (Reference)
#> ST-T wave Abnormality
#> Left Ventricular Hypertrophy
#> fbs:
#> Fasting Blood Sugar <= 120 (Reference)
#> Fasting Blood Sugar >120#>
#> Call: glm(formula = disease ~ cp + age + sex + restecg + fbs, family = binomial(),
#> data = heart_disease)
#>
#> Coefficients:
#> (Intercept) cpNon-anginal Pain
#> -3.97563 -2.30854
#> cpAtypical Angina cpTypical Angina
#> -2.38883 -2.31736
#> age sexMale
#> 0.06316 1.77141
#> restecgST-T wave Abnormality restecgLeft Ventricular Hypertrophy
#> 1.62429 0.50215
#> fbsFasting Blood Sugar >120
#> 0.08444
#>
#> Degrees of Freedom: 296 Total (i.e. Null); 288 Residual
#> Null Deviance: 409.9
#> Residual Deviance: 286.6 AIC: 304.6\[ lo(disease) = -3.0512 + 0.0529 (age) \]
Interpretation
\[ lo(disease) = -3.0512 + 0.0529 (age) \]
As age increases by 1 year, the odds of having heart disease increases by a factor of 1.054.
Prediction with Models
Modeling Binary Outcomes
Logistic Regression
Logistic Regression in R
Prediction with Models
Working with probabilities
As you can see, working with odds may be unintuitive for the average person. It will be better to predict the probability and display those results to individuals.
Predicting Probability
\[ \hat P\left(Y = 1\right) = \frac{e^{\hat\beta_0 + \hat\beta_1X}}{1 + e^{\hat\beta_0 + \hat\beta_1X}} \]
Prediction in R
Example 1
Predict the probability of having heart disease for a patient who is 40 years old.
Example 2
Predict the probability of having heart disease for a patient who is 55 years old.
Example 3
Predict the probability of having heart disease for a patient who is 70 years old.
Example
| Age | 40 | 55 | 70 |
|---|---|---|---|
| Probabiltiy | 28.2% | 46.5% | 65.8% |
m201.inqs.info/lectures/6